Virus-induced gene silencing and virus-induced flowering in strawberry (Fragaria × ananassa) using apple latent spherical virus vectors
Strawberry (Fragaria × ananassa) is a popular crop worldwide with production exceeding four million tons per year1. Despite the long cultivation history of strawberries from the fiteenth century, strawberry breeding remains a major concern of farmers and researchers with regard to a variety of traits such as fruit quality (taste), fruit colour, fruit firmness, fruit storability, ascorbic acid content, everblooming habit, disease resistance and heat/cold resistance2. Changes in these agronomically important traits are expected to be caused by mutations in genomic DNA. For example, the everblooming (continuous flowering) trait of woodland strawberry (Fragaria vesca) is caused by a 2 bp deletion in the coding region of the KSN gene3, a strawberry homologue of the Terminal Flower 1 (TFL1) gene. The genetic mapping approach has also identified the FaOMT gene mutation as the regulator of variation in the release of mesifurane, one of the volatiles of strawberry fruit, based on complete co-segregation of the identified 30-bp mutation in the FaOMT promoter4.
Specific genetic mutations causing changes in other agronomically important traits in strawberry remain mostly unknown, although candidate genes are being suggested through experimental efforts such as gene expression analyses5,6,7,8. The release of genomic sequences and reports of thousands of DNA markers to detect polymorphisms on the chromosomes are also expected to greatly accelerate both forward and reverse genetic studies of strawberry9. Such knowledge with regard to agronomically important genes and their mutations both informs basic plant biology and accelerates strawberry breeding based on DNA information10.
Following identification of agronomically important or candidate genes, their characterization by genetic overexpression or suppression represents a standard strategy for confirmation of their functions. The protocol for genetic transformation of strawberry by Agrobacterium tumefaciens was first established in 199011,12. Whereas the transformation efficiency is generally around 5%, the efficiency may be increased up to 100%13,14,15,16, although transformation rate appears to depend on the strawberry cultivar. Specifically, 100% transformation efficiency was achieved in an everbearing (day-neutral) cultivar ‘Calypso’, using the super-virulent Agrobacterium strain AGL014.
Transient expression/suppression by infiltration of Agrobacterium into strawberry fruits has also been utilized for rapid analysis of gene functions. In this case, Agrobacterium causes overexpression or RNA interference of the target gene17,18, depending on the nucleotide sequence introduced into the transfer DNA region of the binary plasmid. The target strawberry gene may also be suppressed via virus-induced gene silencing (VIGS) using tobacco rattle virus (TRV) vectors19,20. Considering that genetic transformation typically requires 15 months from the start of the experiments to the production of strawberry fruits18, these transient systems enable much faster estimation of gene function than stable transformation, although the genes are expressed/suppressed only in sections within strawberry fruits (more strictly, receptacles). Alternatively, genes can be expressed/suppressed in whole strawberry fruits when Agrobacteriumis repeatedly injected into fruits at least three times18. Such transient systems are currently utilized for the analysis of gene functions with regard to strawberry fruit phenotypes, such as pigmentation, aroma generation, ripening and disease resistance21. Genes can also be transiently expressed in strawberry leaves by infiltration of Agrobacterium for the analysis of their functions22.
Once agronomically important mutations in the genome are identified, they can be combined by cross-breeding and DNA marker selection. However, the long generation time of crops often constitutes a major problem for efficient cross-breeding. Although the genomes of vegetatively propagated cultivars/crops such as apple and pear remain genetically heterozygous, short generation time is also important when the crop genome is homogenized for establishment of seed-propagated genetically homozygous cultivars/crops and F1 hybrid cultivars/crops, such as rice and maize. Thus, in addition to controlling growth conditions such as cultivation in greenhouse or incubator, high CO2levels, tiller removal, paclobutrazol treatment, grafting on rootstock, and embryo rescue23,24,25,26, transgenic expression of the leafy or flowering locus T (FT) gene or suppression of the TFL1 gene also comprise important techniques for inducing early flowering and reducing generation time25,27,28.
Virus-induced flowering (VIF) can also be effective for reducing the generation time of crops. In VIF, crops are infected with RNA virus vectors expressing an FT gene to induce early flowering29. An advantage of VIF is that the genomic DNA of crops is not transformed, and the infected transgenic virus is rarely carried to the progeny (next-generation) plants. In addition, virus infection does not depend on the specific crop cultivar in many cases. Rather, infectivity of virus vectors depends on the host range of viruses; thus, zucchini yellow mosaic virus has been used for the expression of Arabidopsis thaliana FT (AtFT) in squash/pumpkin (Cucurbita moschata)30, cotton leaf crumple virus was used for AtFT expression in cotton31 and citrus leaf blotch virus was used for AtFT or citrus FT expression in citrus plants32 for the successful induction of early flowering. As apple latent spherical virus (ALSV) exhibits a relatively wide host range, in previous studies we used an ALSV vector for the expression of AtFT or other FT genes such as a gentian FT to induce early flowering in A. thaliana, tobacco (Nicotiana) plants, soybean, apple, petunia, lisianthus (eustoma) and gentian33,34,35,36,37. The progeny plants of cross-bred gentian generated by using ALSV-based VIF were confirmed for elimination of the virus, cleared governmental review and started to be grown in an open field in Hachimantai City of Japan for selection of a new cultivar. This appears to be the first example of the application of any New Plant Breeding Technique in Japan.
Again, ALSV enjoys advantages as a virus vector: ALSV has a wide host range, latently infects most plant species (without viral symptoms) and evenly infects the upper leaves. ALSV may provide novel high-throughput methods for estimation of the functions of strawberry genes, and promotion of cross-breeding by shortening the generation time of strawberry. To examine the utility of this technique, in the present study we inoculated ALSV vectors to a commercially important cultivar of strawberry. As a result, the abilities of the ALSV vector to successfully infect strawberry seedlings and to spread to every organ of the shoot, as well as to induce VIGS and VIF in strawberry plants, were ascertained.
Results
Conditions of ALSV inoculation
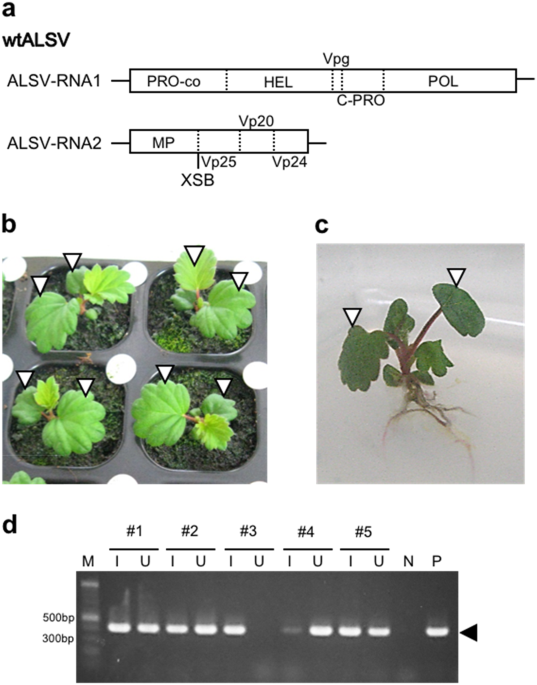
As ALSV vectors have not yet been tested in strawberry, we first aimed to identify a successful condition of ALSV inoculation to strawberry. ALSV consists of RNA1 and RNA2 genomes, with each genome encoding a single polyprotein. For comparison of inoculation conditions, ALSV vector without any insertion of external sequence (wild-type; wtALSV) was used (Fig. 1a). Because ALSV does not necessarily infect horticultural plants easily, ALSV is usually inoculated as viral RNA instead of via rub inoculation or agroinfiltration. For example, gold particles coated with viral RNA are bombarded by gene guns to cotyledons of germinating seeds in the case of apple33,36, or to true leaves of juvenile seedlings in the case of gentian.
Strawberry (Fragaria × ananassa) is a popular crop worldwide with production exceeding four million tons per year1. Despite the long cultivation history of strawberries from the fiteenth century, strawberry breeding remains a major concern of farmers and researchers with regard to a variety of traits such as fruit quality (taste), fruit colour, fruit firmness, fruit storability, ascorbic acid content, everblooming habit, disease resistance and heat/cold resistance2. Changes in these agronomically important traits are expected to be caused by mutations in genomic DNA. For example, the everblooming (continuous flowering) trait of woodland strawberry (Fragaria vesca) is caused by a 2 bp deletion in the coding region of the KSN gene3, a strawberry homologue of the Terminal Flower 1 (TFL1) gene. The genetic mapping approach has also identified the FaOMT gene mutation as the regulator of variation in the release of mesifurane, one of the volatiles of strawberry fruit, based on complete co-segregation of the identified 30-bp mutation in the FaOMT promoter4.
Specific genetic mutations causing changes in other agronomically important traits in strawberry remain mostly unknown, although candidate genes are being suggested through experimental efforts such as gene expression analyses5,6,7,8. The release of genomic sequences and reports of thousands of DNA markers to detect polymorphisms on the chromosomes are also expected to greatly accelerate both forward and reverse genetic studies of strawberry9. Such knowledge with regard to agronomically important genes and their mutations both informs basic plant biology and accelerates strawberry breeding based on DNA information10.
Following identification of agronomically important or candidate genes, their characterization by genetic overexpression or suppression represents a standard strategy for confirmation of their functions. The protocol for genetic transformation of strawberry by Agrobacterium tumefaciens was first established in 199011,12. Whereas the transformation efficiency is generally around 5%, the efficiency may be increased up to 100%13,14,15,16, although transformation rate appears to depend on the strawberry cultivar. Specifically, 100% transformation efficiency was achieved in an everbearing (day-neutral) cultivar ‘Calypso’, using the super-virulent Agrobacterium strain AGL014.
Transient expression/suppression by infiltration of Agrobacterium into strawberry fruits has also been utilized for rapid analysis of gene functions. In this case, Agrobacterium causes overexpression or RNA interference of the target gene17,18, depending on the nucleotide sequence introduced into the transfer DNA region of the binary plasmid. The target strawberry gene may also be suppressed via virus-induced gene silencing (VIGS) using tobacco rattle virus (TRV) vectors19,20. Considering that genetic transformation typically requires 15 months from the start of the experiments to the production of strawberry fruits18, these transient systems enable much faster estimation of gene function than stable transformation, although the genes are expressed/suppressed only in sections within strawberry fruits (more strictly, receptacles). Alternatively, genes can be expressed/suppressed in whole strawberry fruits when Agrobacteriumis repeatedly injected into fruits at least three times18. Such transient systems are currently utilized for the analysis of gene functions with regard to strawberry fruit phenotypes, such as pigmentation, aroma generation, ripening and disease resistance21. Genes can also be transiently expressed in strawberry leaves by infiltration of Agrobacterium for the analysis of their functions22.
Once agronomically important mutations in the genome are identified, they can be combined by cross-breeding and DNA marker selection. However, the long generation time of crops often constitutes a major problem for efficient cross-breeding. Although the genomes of vegetatively propagated cultivars/crops such as apple and pear remain genetically heterozygous, short generation time is also important when the crop genome is homogenized for establishment of seed-propagated genetically homozygous cultivars/crops and F1 hybrid cultivars/crops, such as rice and maize. Thus, in addition to controlling growth conditions such as cultivation in greenhouse or incubator, high CO2levels, tiller removal, paclobutrazol treatment, grafting on rootstock, and embryo rescue23,24,25,26, transgenic expression of the leafy or flowering locus T (FT) gene or suppression of the TFL1 gene also comprise important techniques for inducing early flowering and reducing generation time25,27,28.
Virus-induced flowering (VIF) can also be effective for reducing the generation time of crops. In VIF, crops are infected with RNA virus vectors expressing an FT gene to induce early flowering29. An advantage of VIF is that the genomic DNA of crops is not transformed, and the infected transgenic virus is rarely carried to the progeny (next-generation) plants. In addition, virus infection does not depend on the specific crop cultivar in many cases. Rather, infectivity of virus vectors depends on the host range of viruses; thus, zucchini yellow mosaic virus has been used for the expression of Arabidopsis thaliana FT (AtFT) in squash/pumpkin (Cucurbita moschata)30, cotton leaf crumple virus was used for AtFT expression in cotton31 and citrus leaf blotch virus was used for AtFT or citrus FT expression in citrus plants32 for the successful induction of early flowering. As apple latent spherical virus (ALSV) exhibits a relatively wide host range, in previous studies we used an ALSV vector for the expression of AtFT or other FT genes such as a gentian FT to induce early flowering in A. thaliana, tobacco (Nicotiana) plants, soybean, apple, petunia, lisianthus (eustoma) and gentian33,34,35,36,37. The progeny plants of cross-bred gentian generated by using ALSV-based VIF were confirmed for elimination of the virus, cleared governmental review and started to be grown in an open field in Hachimantai City of Japan for selection of a new cultivar. This appears to be the first example of the application of any New Plant Breeding Technique in Japan.
Again, ALSV enjoys advantages as a virus vector: ALSV has a wide host range, latently infects most plant species (without viral symptoms) and evenly infects the upper leaves. ALSV may provide novel high-throughput methods for estimation of the functions of strawberry genes, and promotion of cross-breeding by shortening the generation time of strawberry. To examine the utility of this technique, in the present study we inoculated ALSV vectors to a commercially important cultivar of strawberry. As a result, the abilities of the ALSV vector to successfully infect strawberry seedlings and to spread to every organ of the shoot, as well as to induce VIGS and VIF in strawberry plants, were ascertained.
Results
Conditions of ALSV inoculation
As ALSV vectors have not yet been tested in strawberry, we first aimed to identify a successful condition of ALSV inoculation to strawberry. ALSV consists of RNA1 and RNA2 genomes, with each genome encoding a single polyprotein. For comparison of inoculation conditions, ALSV vector without any insertion of external sequence (wild-type; wtALSV) was used (Fig. 1a). Because ALSV does not necessarily infect horticultural plants easily, ALSV is usually inoculated as viral RNA instead of via rub inoculation or agroinfiltration. For example, gold particles coated with viral RNA are bombarded by gene guns to cotyledons of germinating seeds in the case of apple33,36, or to true leaves of juvenile seedlings in the case of gentian.

Comments
Post a Comment
Please comment on this blog-